In certain areas of the world it is difficult to find medical staff to make the diagnosis of diseases such as malaria. The World Health Organization (WHO) announced 445,000 malaria deaths in 2016. Much of this is especially true in remote areas of Africa. On the other hand, there are rapid diagnostic tests to diagnose malaria but these are not as reliable as microscopy that remains the mainstay of parasite-based diagnosis in most large health clinics. There are several diseases, not only malaria, which can be diagnosed through a blood sample and a microscope, it is the case of filarial parasites.
In this project we have built a model to detect malaria in human blood images. The idea is to have a cheap and reliable system that can deliver results without the need for highly specialized staff. In this way one could increase the diagnostic in useful time reducing the malaria mortality rate.
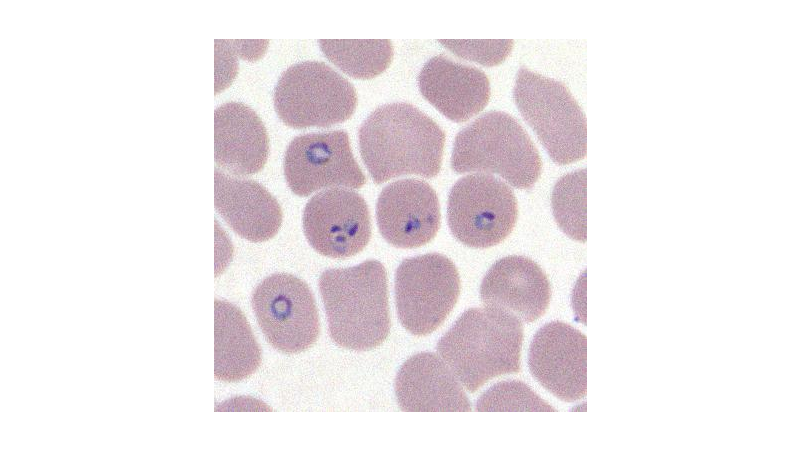
These images are formed, mainly, by erythrocytes, leukocytes and malaria parasites.
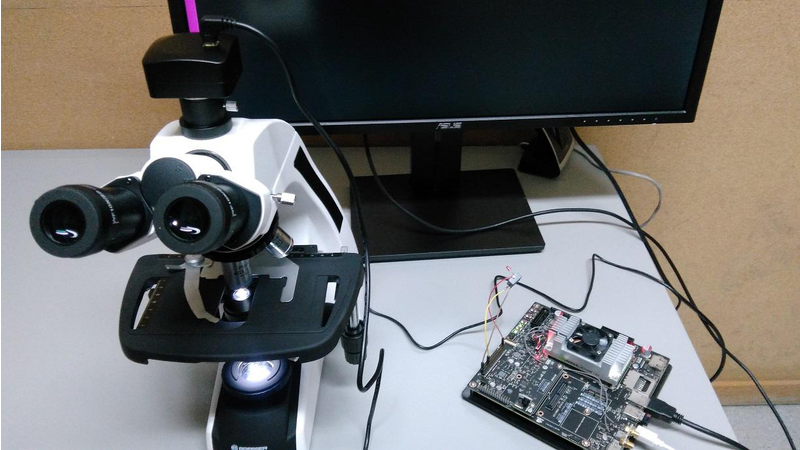
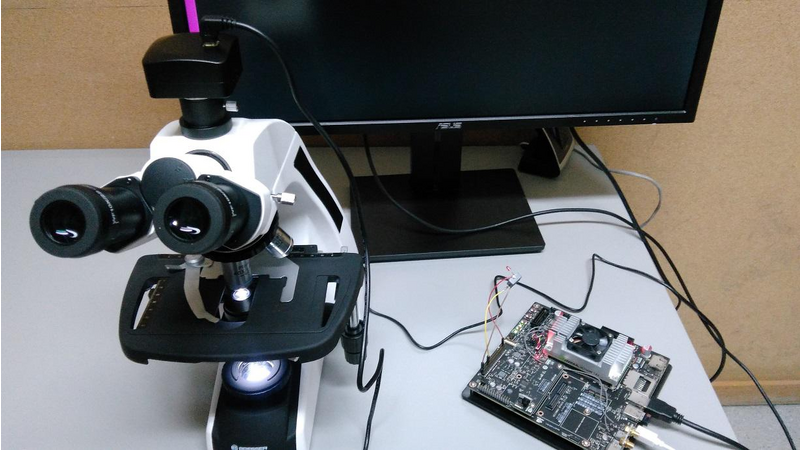
The system consists of three fundamental parts: The Jetson TX2, a digital camera and a microscope.
The Jetson receives the microscope color image from the digital camera and analyzes it according the deep learning model. That model was built using the Digits 6 framework, specifically, using a DetectNet architecture. The training set is formed by (roughly) 2000 regions of malaria parasites, at different growing stages, annotated by our pathologist using the KITTI metadata format. The negative examples, 4000, are extracted from the blood smear, some of them with the disease and some of them without the disease. The negative examples correspond to a non-parasite regions. The images set was split to get the validation and test sets.
During the training the mAP(val) reached the 40.8 at the epoch 310.
In order to increase the robustness to the noise of the camera, an output stage was implemented. It runs over several frames evaluating the coherence of the boxes pointed out by DetectNet. This consistency is evaluated using the Intersection over Union (IoU) metric where each box of a frame is compared to the corresponding boxes along the frames. Thus, malaria region detections have to remain consistent for several frames to be announced the malaria detection with a high degree of certainty.
The Jetson GPIO port is used to drive a LED and a buzzer when a malaria detection is performed.
The original detect-camera.cpp source code was adapted to this solution in manner to have the GPIO control and the IoU approach integrated in the inference engine.
Results
The results are showing a good performance of the system because in 50 images of malaria and 50 images of non-malaria the system was successful in the classification reaching a rate of 100% (no errors).
Nevertheless, some malaria stages could be mismatched to some leukocytes. Thus, in order to reduce this error we need to increase the number of training images.
Instructions
1 – A glass slide is prepared with a blood smear. The slide is stained (Wright-Giemsa stained) to distinguish the cells from each other;
2 – Put the glass slide on the microscope and make some adjustments to the microscope focus;
3 – Turn on Jetson and see the answer (Malaria or non-Malaria) looking to the external LED and hearing the buzzer;
4 – Move the slide adjusting the microscope table for a new optical field (xy position).
5 – Get a new answer from Jetson to confirm the previous one.
Future Work
The model, eventually, could be improved with new images of malaria.
New diseases can be learnt by the model.
A simpler and portable microscope can be built reducing the final price and allowing to put all together in a box (Jetson + camera + microscope).


Comments (0)