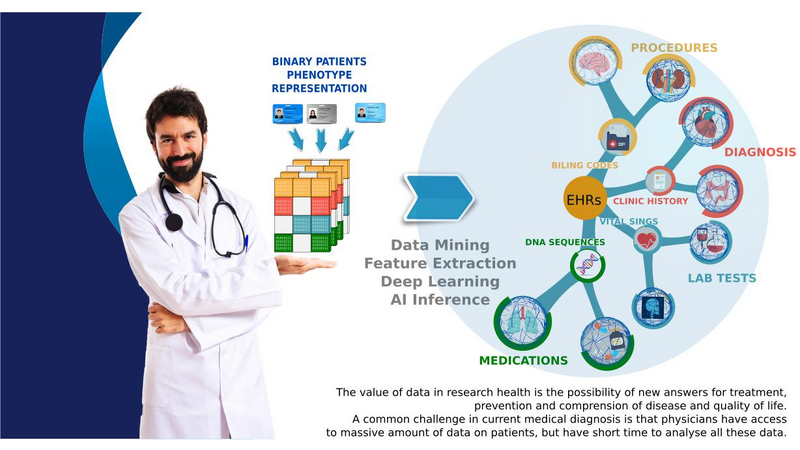
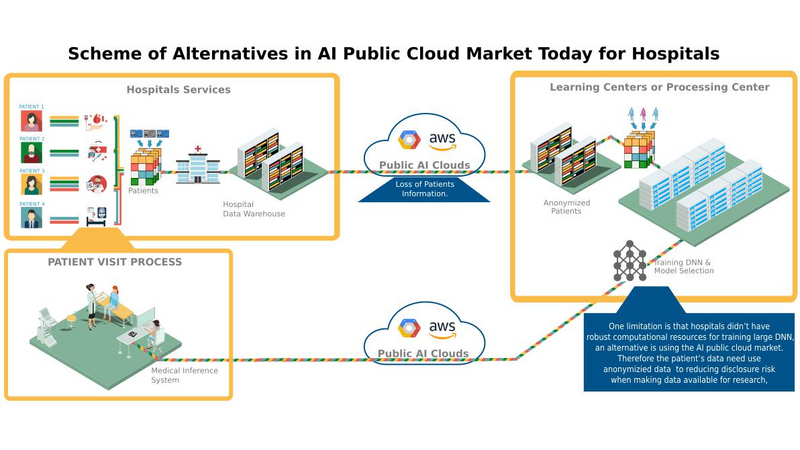
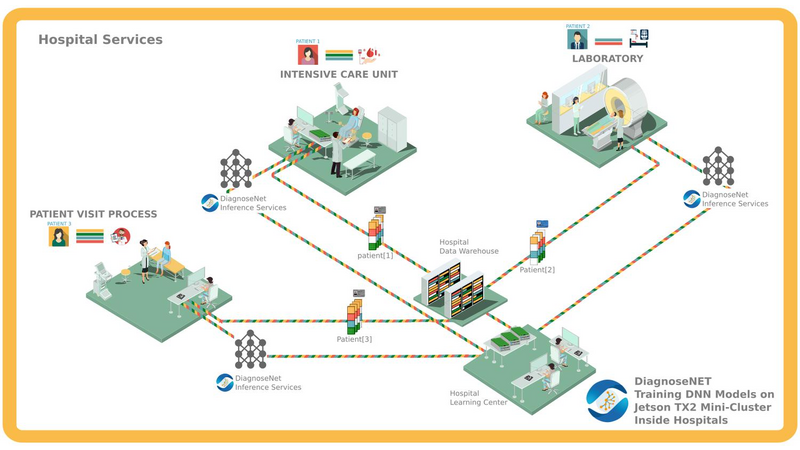
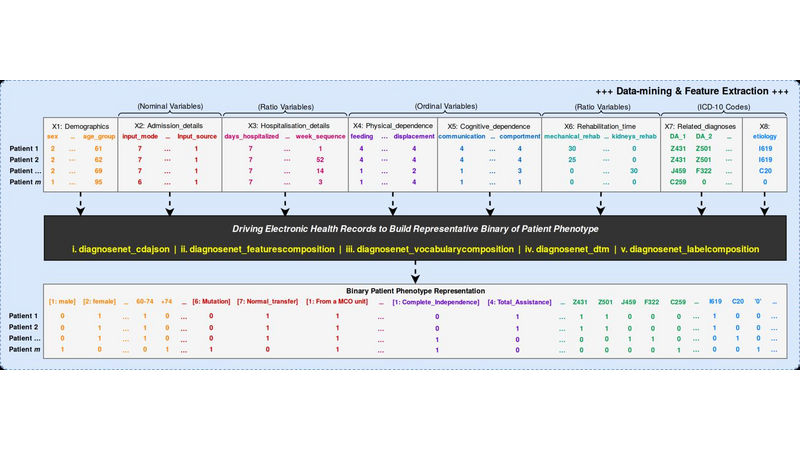
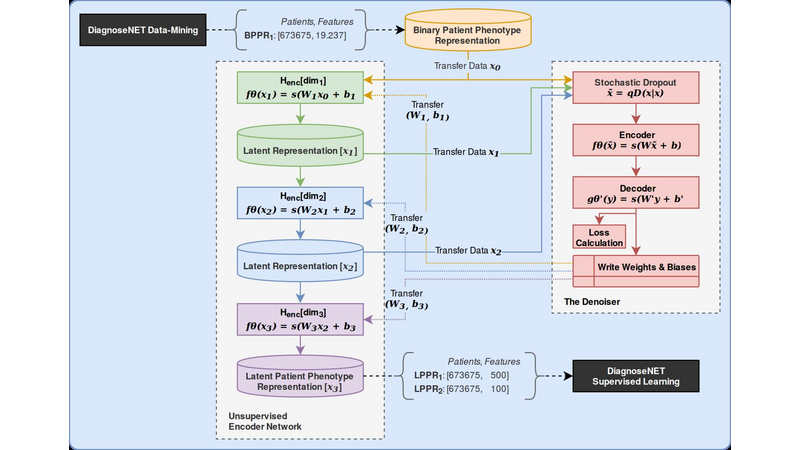
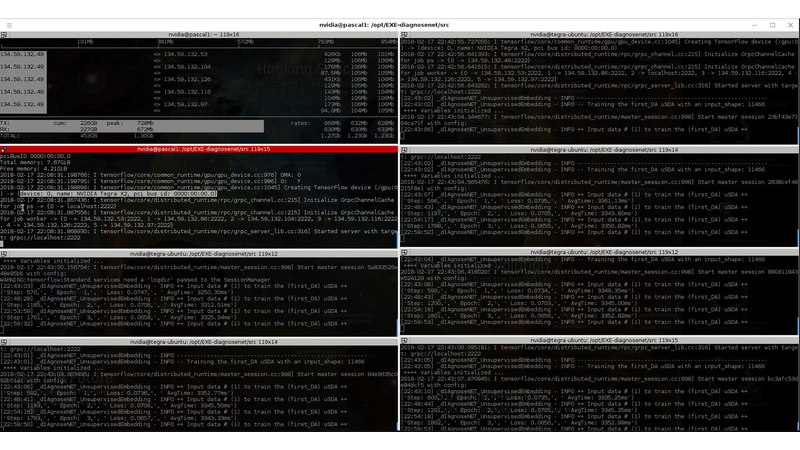
DiagnoseNet: Framework for Training Dee Neural Networks on Mini-Cluster Jetson TX2 Inside the Hositals<>The value of data in research health is the ossiity of new answers for treatment, revention and comrehension of disease and quaty of fe. A common challenge in current medical diagnosis is that hysicians have access to massive amount of data on atients, ut have the short time to analyse all these data. Choi et al. (2015)One mitation is that hositals didn’t have roust comutational resources for training large dee neural networks, an alternative is using the AI ic cloud market. Therefore the atient’s data need use anonymizing data to reducing disclosure risk when making data availale for research, this rocedure cod lose the truthfness at the record level and cause wrong mining rests. Nguyen TA. et al (2017).<> We are uilding a framework called DiagnoseNET for training fl Dee Neural Networks (DNN) on&ns; Mini-Cluster Inside the Hositals. Therefore, this can e used as a learning centre with minimal infrastructure requirements, low ower consumtion and rings the oortunity to use a dataset with more atient’s features.<> DiagnoseNET that rovides three high-level features:<> a framework to uild fl Dee Neural Networks (DNN) workflow;<> a distriuted rocessing training DNN on Mini-Cluster Jetson TX2;<> and an energy-monitoring to for workload characterization. <> <> DiagnoseNet-01 imlements the unsuervised dee learning method resented y Miotto et al. (2016) which goal is to create a new lower dimensional atient reresentation for cnical redictive modelng. This workflow is divided into three stages:<><> DiagnoseNet Data-Mining<> Is a feature extraction API that use a dynamic features comosition with their resective vocaary to uild a inary atient henotye reresentation in a 'document-term sarse matrix' from ICU dataset.<> DiagnoseNet Unsuervised Emedding<> This API re-training three Denoising Autoencoder (DAE) to get each latent reresentation and sends them as inut to the next DAE resectively. The Encoder Network uses a clean BPPR as inut for maing with the reviously trained weights to otain a new Latent Patient Phenotye Reresentation (LPPR).<> DiagnoseNet Suervised Learning<> To train the BPPR and LPPR y random forest algorithm for classification and rediction of medical inatient states at intensive care unit in PACA region.<><> <> <> <> Predict the Medical Future of Patients from EHRs<>:Driving Electronic Health Records (EHR) to uild a general atient henotye reresentation, resents key challenges in ig data, dee learning and arallel rocessing as the high energy cost for training models until finding the otimal model generazation-accuracy. The general dataset is comosed of diagnosis-related grou reresented in oject form; such as ICD-10 codes, CCAM codes and other codes estashed y the agency ATIH and generated y the system PMSI for the Intensive Care Unit (ICU) and Cnical Oerations (MCO) with activities of hositals in PACA and the activities of residents PACA hositaze in another region.&ns;&ns; <> As a case of study, we have used an Intensive Care Unit (ICU) dataset with an average of 785,801 Inatients records y year, to uild a general urose inatient henotye reresentation for general acative medical targets, such as: redict the ‘Major Cnical Category’ from inatients features recorded in hosital admission and cnical attention. <> Preminary Study to Predict the Cnical Major Category as First Medical Target:<><> Stage 1:<> We resent two ways to derive a BPPR&ns; selecting different Inatient features groued y high-level entities as was shown at ICU data clection scheme (i.e X1_Demograhics: {sexe: [], age: [], ...}), as rest, we got two different BPPR in length of features and disk size, such as BPPR1: {Features: 11094}, {Patients: 99999} {disk: 3.2 GB}, BPPR2: {Features: 14515}, {Patients: 99999} {disk: 4.1 GB}. As an exeriment are using 12% of ICU data y the 2008 year.&ns; <> Stage 2: <>We use two grous of layer dimensions with the same hyerarameters (i.e. Relu, Adadelta, Learning rate: 0.001, atch size: 32, num. eoch: 50, ...) to change the sace and encode the length and disk size of the revious BPPR1 to LPPR11: {Features: 500}, {Patients: 99999} {disk: 255 MB} and LPPR12: {Features: 100}, {Patients: 99999} {disk: 7 MB}. As an exeriment, each revious BPPR was divided into 9 atch files.&ns;&ns; <> Stage 3: <>We are imlemented Random Forest Classifier (RFC) to redict the Cnical Major Category (CMC) laels that were resented in the first medical target. As an exeriment we set the same RFC arameters (i.e. estimators: 100, max_features: 'sqrt', ...) to comare the recision in terms of (AUC-ROC) etween each revious BPPR1 and their two LPPR11 and LPPR12, the rests were 0.792, 0.835, 0.83, resectively, good recision statistics.Preminary Rests<><> Use the unsuervised emedding stage to create a new lower dimensional atient reresentation, reduces the numer of sarse features to classify at stage 3. In which, the execution time for training is minimized y 41% with regard to BPPR and the recision to classify the first medical target is almost equal.<> <> The unsuervised emedding stage reresents 64% of the fl DNN workflow execution time. To imrove this rocessing stage it is necessary to aralleze the USDA to exloit the comutational resources and using alternative latforms (i.e. emedding systems) to minimize the ower consumtion.<> <> <><> Distriuted Unsuervised Emedding<><> We are uilding a Mini-cluster Jetson TX2, starting with 8 Jetson oards and we have lanned to work to 12, 24 oards to evaluate their erformance. We started with the imlementation of the distriuted rocessing at stage 2 from the fl DNN workflow, that we called DiagnoseNET unsuervised emedding. To exloit the arallel and distriuted rocessing we selected the data arallesm, where we st the BPPR in several mini-atch and assigned each grou to one jetson oard. The first technique imlemented is the synchronous training, were all recas task read the same values for the current arameters, comute gradients in arallel, and theirs aly together.&ns; Jeffrey Dean et al. (2012)<> <> <> References<><> [1] Edward Choi, Mohammad Taha Bahadori, and Jimeng Sun. Doctor AI: Predicting CnicalEvents via Recurrent Neural Networks. CoRR, as1511.05942, 2015. URL htt:arxiv.orgas1511.05942;htt:dl.orgrecijournalscorrChoiBS15.<> [2] Nguyen TA., Le-Khac NA., Kechadi MT. (2017) Privacy-Aware Data Analysis Middleware for Data-Driven EHR Systems. In: Dang T., Wagner R., Küng J., Thoai N., Takizawa M., Neuhd E. (eds) Future Data and Security Engineering. FDSE 2017. Lecture Notes in Comuter Science, v 10646. Sringer, Cham.<> [3] Riccardo Miotto, Li Li, Brian A. Kidd, and Joel T. Dudley. Dee Patient: An UnsuervisedReresentation to Predict the Future of Patients from the Electronic Health Records.SCIENTIFIC REPORTS, 2016. URL htt:www.nature.comarticlessre26094.<> [4] Jeffrey Dean, Greg S. Corrado, Rajat Monga, Kai Chen, Matthieu Devin, Quoc V. Le, Mark Z. Mao, Marc'Aureo Ranzato, Andrew Senior, Pa Tucker, Ke Yang, and Andrew Y. Ng. Large Scale Distriuted Dee Networks. NIPS 2012: Neural Information Processing Systems.<> <>
Let’s talk
Log in
Not a candidate? Sign up as an employer
Reset your password
Remember your password? Log in Log in for business
Create an employer account
Sign up for free.
Select the best plan to publish job ofers & challenges.
Not an employer? Sign up as a candidate




Comments (3)
JP
Julian Pelaez-Restrepo
SM
Sergio Machuca
1
1825515